9 PCA Plots
9.1 Introduction
This tutorial is based on a nice blog post by Claus Wilke: https://clauswilke.com/blog/2020/09/07/pca-tidyverse-style/
Functions from the Tidyverse packages (dplyr , tidyr, ggplot2) and broom will be used in this tutorial. As example data a lipidomics dataset is imported that contains the lipid concentrations as columns and with one additional column indicating the experimental group
| SubjectID | Group | CE 16:0 | CE 17:0 | CE 17:1 | CE 18:0 | CE 18:1 | CE 18:2 | CE 20:1 | CE 20:2 | CE 20:3 | CE 20:4 | CE 20:5 | CE 22:4 | CE 22:6 | CE 24:4 | Cer d18:0/16:0 | Cer d18:0/18:0 | Cer d18:0/20:0 | Cer d18:0/22:0 | Cer d18:0/24:1 | Cer d18:1/16:0 | Cer d18:1/18:0 | Cer d18:1/20:0 | Cer d18:1/22:0 | Cer d18:1/24:0 | Cer d18:1/24:1 | Cer d18:1/25:0 | Cer d18:1/26:0 | Cer d18:2/22:0 | Cer d18:2/24:1 | DG 14:0_18:1 | DG 16:0_16:0 | DG 16:0_16:1 | DG 16:0_20:3 | DG 16:0_20:4 | DG 16:0_22:6 | DG 16:1_18:1 | DG 18:0_18:1 | DG 18:0_18:2 | DG 18:0_20:4 | DG 18:1_18:1 | DG 18:1_18:2 | DG 18:1_18:3 | DG 18:1_20:3 | DG 18:1_20:4 | DG 18:2_18:2 | DG 18:2_20:3 | DG 18:2_20:4 | GM3 d18:1/16:0 | GM3 d18:1/18:0 | GM3 d18:1/20:0 | GM3 d18:1/22:0 | Hex2Cer d18:1/16:0 | Hex2Cer d18:1/18:0 | Hex2Cer d18:1/22:0 | Hex2Cer d18:1/24:0 | Hex2Cer d18:1/24:1 | Hex3Cer d18:1/16:0 | HexCer d18:1/18:0 | HexCer d18:1/20:0 | HexCer d18:1/22:0 | HexCer d18:1/24:0 | HexCer d18:1/24:1 | LPC 14:0 | LPC 15:0 | LPC 16:1 | LPC 17:0 | LPC 17:1 | LPC 18:0 | LPC 18:1 | LPC 18:2 | LPC 18:3 | LPC 19:0 | LPC 19:1 | LPC 20:0 | LPC 20:1 | LPC 20:2 | LPC 20:3 | LPC 20:4 | LPC 20:5 | LPC 22:0 | LPC 22:1 | LPC 22:5 | LPC 22:6 | LPC 24:0 | LPC O-16:0 | LPC O-18:0 | LPC O-18:1 | LPC O-20:0 | LPC O-20:1 | LPC O-22:0 | LPC O-22:1 | LPC P-16:0 | LPC P-18:0 | LPC P-18:1 | LPC P-20:0 | LPE 16:0 | LPE 18:0 | LPE 18:1 | LPE 18:2 | LPE 20:4 | LPE 22:6 | LPE P-16:0 | LPE P-18:0 | LPE P-18:1 | LPI 20:4 | LPS 16:0 | LPS 18:0 | LPS 18:1 | LPS 18:2 | PC 31:0 | PC 31:1 | PC 32:0 | PC 32:1 | PC 32:2 | PC 32:3 | PC 33:1 | PC 33:2 | PC 34:0 | PC 34:2 | PC 34:3 | PC 34:4 | PC 34:5 | PC 35:1 | PC 35:2 | PC 35:3 | PC 35:4 | PC 35:5 | PC 36:1 | PC 36:2 | PC 36:3 | PC 36:4 a | PC 36:4 b | PC 36:5 | PC 36:6 | PC 36:7 | PC 37:4 | PC 37:5 | PC 37:6 | PC 38:3 | PC 38:6 | PC 38:7 | PC 38:8 | PC 39:5 | PC 39:6 | PC 39:7 | PC 40:6 | PC 40:7 | PC 40:8 | PC O-32:0 | PC O-32:1 | PC O-32:2 | PC O-34:1 | PC O-34:2 | PC O-36:1 | PC O-36:2 | PC O-36:4 | PC O-36:5 | PC O-38:4 | PC O-38:5 | PC O-38:6 | PC O-40:5 | PC O-40:7 | PC P-32:0 | PC P-32:1 | PC P-34:1 | PC P-34:2 | PC P-36:2 | PC P-36:4 | PC P-36:5 | PC P-38:4 | PC P-38:5 | PC P-40:5 | PC P-40:6 | PE 32:1 | PE 34:0 | PE 34:1 | PE 34:2 | PE 34:3 | PE 35:1 | PE 35:2 | PE 36:1 | PE 36:2 | PE 36:4 a | PE 36:4 b | PE 36:5 | PE 38:3 | PE 38:4 | PE 38:5 | PE 38:6 | PE 40:4 | PE 40:6 | PE 40:7 | PE O-34:1 | PE O-34:2 | PE O-36:2 | PE O-36:3 | PE O-38:4 | PE O-38:5 | PE O-40:5 | PE O-40:7 | PE P-16:0/18:1 | PE P-16:0/18:2 | PE P-16:0/20:4 | PE P-16:0/22:5 | PE P-16:0/22:6 | PE P-18:0/18:1 | PE P-18:0/18:2 | PE P-18:0/20:4 | PE P-18:0/22:5 | PE P-18:0/22:6 | PE P-20:0/20:4 | PG 34:1 | PG 36:1 | PG 36:2 | PI 32:0 | PI 32:1 | PI 34:1 | PI 34:2 | PI 35:1 | PI 35:2 | PI 36:2 | PI 36:4 | PI 36:5 | PI 37:4 | PI 38:4 | PI 38:5 | PI 38:6 | PI 40:6 | S1P d16:1 | S1P d17:1 | S1P d18:0 | S1P d18:1 | S1P d18:2 | S1P d19:1 | S1P d20:1 | SM 30:1 | SM 31:1 | SM 32:0 | SM 32:1 | SM 32:2 | SM 33:1 | SM 34:1 | SM 34:2 | SM 34:3 | SM 35:1 | SM 35:2 | SM 36:1 | SM 36:2 | SM 36:3 | SM 38:1 | SM 38:2 | SM 38:3 | SM 39:1 | SM 40:1 | SM 40:2 | SM 41:1 | SM 42:1 | SM 42:2 | SM 43:1 | SM 44:1 | SM 44:2 | TG 48:0 | TG 48:1 | TG 48:2 | TG 48:3 | TG 49:1 | TG 50:0 | TG 50:1 | TG 50:2 | TG 50:3 | TG 50:4 | TG 51:0 | TG 51:1 | TG 51:2 | TG 52:1 | TG 52:2 | TG 52:3 | TG 52:4 | TG 53:2 | TG 54:1 | TG 54:2 | TG 54:3 | TG 54:4 | TG 54:5 | TG 54:6 | TG 56:6 | TG 56:8 | TG 58:8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HB_01 | Healthy | 36.82 | 1.02 | 1.34 | 12.34 | 573.83 | 4783.39 | 0.38 | 5.82 | 116.52 | 4123.15 | 44.71 | 12.74 | 33.72 | 0.30 | 0.02 | 0.03 | 0.02 | 0.05 | 0.85 | 0.20 | 0.29 | 0.21 | 1.44 | 2.38 | 2.00 | 0.07 | 0.03 | 0.12 | 0.17 | 0.49 | 1.11 | 1.27 | 0.69 | 3.83 | 0.12 | 5.70 | 2.05 | 4.96 | 3.19 | 30.36 | 34.52 | 3.43 | 1.56 | 6.97 | 12.12 | 1.47 | 5.26 | 2.39 | 1.07 | 0.20 | 0.11 | 4.76 | 0.20 | 0.13 | 0.10 | 0.21 | 0.74 | 0.07 | 0.04 | 0.09 | 0.11 | 0.09 | 0.80 | 1.85 | 6.25 | 2.50 | 0.30 | 197.98 | 52.83 | 105.47 | 0.79 | 0.84 | 0.12 | 0.24 | 0.69 | 0.83 | 5.45 | 94.90 | 0.60 | 0.06 | 0.03 | 6.66 | 1.87 | 0.07 | 1.62 | 0.58 | 1.62 | 0.04 | 0.04 | 0.00 | 0.01 | 0.30 | 0.04 | 0.04 | 0.04 | 1.99 | 4.55 | 2.43 | 2.46 | 1.53 | 0.16 | 0.31 | 0.47 | 0.13 | 0.33 | 0.27 | 0.31 | 0.07 | 0.15 | 0.69 | 0.14 | 18.23 | 20.27 | 4.94 | 0.11 | 3.07 | 8.96 | 6.35 | 725.25 | 56.00 | 2.07 | 0.13 | 10.78 | 17.68 | 4.78 | 3.34 | 0.02 | 140.27 | 397.37 | 195.86 | 63.39 | 347.22 | 21.84 | 0.36 | 0.03 | 36.10 | 5.71 | 0.20 | 75.69 | 83.26 | 1.73 | 0.01 | 8.12 | 0.95 | 0.10 | 31.54 | 7.50 | 4.86 | 5.60 | 2.73 | 0.59 | 15.95 | 5.95 | 0.83 | 4.54 | 61.03 | 2.69 | 26.80 | 44.17 | 2.25 | 7.63 | 1.42 | 2.32 | 0.46 | 4.77 | 8.37 | 2.37 | 16.51 | 0.79 | 9.47 | 7.78 | 1.65 | 0.16 | 5.00 | 0.05 | 0.68 | 1.27 | 0.19 | 0.05 | 0.10 | 0.51 | 3.16 | 0.90 | 2.22 | 0.11 | 0.22 | 5.39 | 2.22 | 2.81 | 0.30 | 0.37 | 0.29 | 0.34 | 0.19 | 1.12 | 0.27 | 7.36 | 3.21 | 3.68 | 0.42 | 0.17 | 0.25 | 0.94 | 1.07 | 0.44 | 0.28 | 0.71 | 3.51 | 0.75 | 0.28 | 0.07 | 0.08 | 0.05 | 0.37 | 0.13 | 0.32 | 3.01 | 5.30 | 0.12 | 0.26 | 9.29 | 11.66 | 0.18 | 0.95 | 74.62 | 6.87 | 0.43 | 0.26 | 0.04 | 0.02 | 0.06 | 0.58 | 0.17 | 0.01 | 0.00 | 0.05 | 0.11 | 0.05 | 1.93 | 0.16 | 4.68 | 389.77 | 8.08 | 0.05 | 4.82 | 0.91 | 24.57 | 31.56 | 0.48 | 8.62 | 3.38 | 0.80 | 6.89 | 17.32 | 10.78 | 10.35 | 7.62 | 33.19 | 0.48 | 0.08 | 0.23 | 5.97 | 7.81 | 21.94 | 6.19 | 1.93 | 3.95 | 28.19 | 53.31 | 54.20 | 1.48 | 0.41 | 1.54 | 4.84 | 16.46 | 70.45 | 29.21 | 95.72 | 1.88 | 1.98 | 23.87 | 113.44 | 24.18 | 48.59 | 16.26 | 10.51 | 7.99 | 0.17 |
| HB_02 | Healthy | 45.57 | 1.11 | 1.28 | 10.16 | 712.27 | 6232.73 | 0.40 | 5.80 | 113.39 | 4415.84 | 57.05 | 14.15 | 42.67 | 0.33 | 0.01 | 0.02 | 0.02 | 0.05 | 1.11 | 0.21 | 0.25 | 0.20 | 1.73 | 2.91 | 2.38 | 0.09 | 0.04 | 0.10 | 0.15 | 0.02 | 0.59 | 0.32 | 0.26 | 1.68 | 0.01 | 1.12 | 1.62 | 3.38 | 1.42 | 5.61 | 6.70 | 0.89 | 0.70 | 5.31 | 2.72 | 0.67 | 3.25 | 3.08 | 1.20 | 0.20 | 0.11 | 7.48 | 0.36 | 0.19 | 0.10 | 0.24 | 0.78 | 0.04 | 0.03 | 0.08 | 0.10 | 0.08 | 1.05 | 1.75 | 6.59 | 2.05 | 0.26 | 153.57 | 52.02 | 108.45 | 0.84 | 0.67 | 0.10 | 0.26 | 0.61 | 0.75 | 3.77 | 74.98 | 0.87 | 0.07 | 0.04 | 5.97 | 1.21 | 0.07 | 1.62 | 0.67 | 1.54 | 0.05 | 0.05 | 0.01 | 0.01 | 0.32 | 0.06 | 0.04 | 0.05 | 2.59 | 4.80 | 2.78 | 3.02 | 1.72 | 0.17 | 0.28 | 0.68 | 0.14 | 0.39 | 0.30 | 0.31 | 0.09 | 0.15 | 0.74 | 0.15 | 17.55 | 30.93 | 6.98 | 0.17 | 3.69 | 8.16 | 6.54 | 614.49 | 71.32 | 2.64 | 0.14 | 11.14 | 15.42 | 4.79 | 4.09 | 0.04 | 115.08 | 457.87 | 190.02 | 53.79 | 333.31 | 31.97 | 0.48 | 0.03 | 28.79 | 6.26 | 0.24 | 57.56 | 88.95 | 2.00 | 0.02 | 10.74 | 0.88 | 0.14 | 23.68 | 7.56 | 4.78 | 4.38 | 2.84 | 0.63 | 16.71 | 6.73 | 1.37 | 6.63 | 65.08 | 3.61 | 31.57 | 55.85 | 2.63 | 10.20 | 1.47 | 2.56 | 0.39 | 5.54 | 9.06 | 3.26 | 23.47 | 1.07 | 9.15 | 11.27 | 1.91 | 0.15 | 4.60 | 0.03 | 0.75 | 1.08 | 0.15 | 0.02 | 0.10 | 0.70 | 2.65 | 0.77 | 2.05 | 0.11 | 0.18 | 7.22 | 1.89 | 1.96 | 0.27 | 0.26 | 0.20 | 0.60 | 0.27 | 2.16 | 0.34 | 9.41 | 4.07 | 5.37 | 0.32 | 0.15 | 0.36 | 1.14 | 2.00 | 0.51 | 0.47 | 1.20 | 4.53 | 1.22 | 0.29 | 0.10 | 0.06 | 0.02 | 0.22 | 0.45 | 0.68 | 4.86 | 8.59 | 0.24 | 0.36 | 19.72 | 11.50 | 0.30 | 1.16 | 85.63 | 6.94 | 0.55 | 0.34 | 0.04 | 0.02 | 0.06 | 0.50 | 0.14 | 0.01 | 0.00 | 0.05 | 0.11 | 0.08 | 2.31 | 0.21 | 4.98 | 397.57 | 9.34 | 0.04 | 4.50 | 0.71 | 23.48 | 41.21 | 0.45 | 9.93 | 2.83 | 0.68 | 6.72 | 15.78 | 10.63 | 10.42 | 8.15 | 33.41 | 0.59 | 0.07 | 0.20 | 5.00 | 1.77 | 5.04 | 1.26 | 1.07 | 2.30 | 12.19 | 15.12 | 9.56 | 0.61 | 0.24 | 0.70 | 1.93 | 11.54 | 29.36 | 8.07 | 16.65 | 0.54 | 1.39 | 16.94 | 33.20 | 5.63 | 7.21 | 2.76 | 9.31 | 3.98 | 0.06 |
| HB_03 | Healthy | 40.64 | 0.87 | 1.58 | 8.20 | 632.30 | 4920.22 | 0.42 | 5.55 | 149.03 | 4034.94 | 51.56 | 17.97 | 61.90 | 0.23 | 0.01 | 0.02 | 0.02 | 0.05 | 1.25 | 0.35 | 0.38 | 0.40 | 2.35 | 3.50 | 3.35 | 0.10 | 0.03 | 0.23 | 0.39 | 0.22 | 0.66 | 0.40 | 0.46 | 2.40 | 0.07 | 1.72 | 0.45 | 0.92 | 1.41 | 9.97 | 11.02 | 1.84 | 1.27 | 8.05 | 4.35 | 1.21 | 5.71 | 2.24 | 0.77 | 0.16 | 0.09 | 3.04 | 0.21 | 0.20 | 0.13 | 0.24 | 0.27 | 0.05 | 0.02 | 0.10 | 0.13 | 0.08 | 1.25 | 1.98 | 8.58 | 2.25 | 0.37 | 156.16 | 54.13 | 91.03 | 0.77 | 0.70 | 0.12 | 0.21 | 0.68 | 0.79 | 5.48 | 77.70 | 0.72 | 0.07 | 0.04 | 9.10 | 1.94 | 0.08 | 1.28 | 0.46 | 1.19 | 0.05 | 0.04 | 0.01 | 0.01 | 0.21 | 0.04 | 0.03 | 0.03 | 2.96 | 5.81 | 2.89 | 3.30 | 1.88 | 0.33 | 0.25 | 0.36 | 0.09 | 0.35 | 0.28 | 0.31 | 0.09 | 0.13 | 0.90 | 0.30 | 13.22 | 31.86 | 9.69 | 0.16 | 4.72 | 8.71 | 5.66 | 635.28 | 84.25 | 3.62 | 0.14 | 11.14 | 14.28 | 5.11 | 4.06 | 0.04 | 119.70 | 446.95 | 186.15 | 53.95 | 314.50 | 31.73 | 0.59 | 0.03 | 29.70 | 6.63 | 0.38 | 85.55 | 112.37 | 2.33 | 0.01 | 8.95 | 1.28 | 0.15 | 39.94 | 11.01 | 5.10 | 4.83 | 2.57 | 0.39 | 13.05 | 4.45 | 0.81 | 3.49 | 44.28 | 2.28 | 22.80 | 45.42 | 1.86 | 6.43 | 1.17 | 1.48 | 0.38 | 4.42 | 5.46 | 2.03 | 14.87 | 0.70 | 6.68 | 7.25 | 1.45 | 0.14 | 11.04 | 0.02 | 1.18 | 2.50 | 0.35 | 0.07 | 0.24 | 1.25 | 4.16 | 1.31 | 3.84 | 0.29 | 0.60 | 11.94 | 3.70 | 6.99 | 0.97 | 0.65 | 0.57 | 0.42 | 0.20 | 0.87 | 0.26 | 6.56 | 3.44 | 4.77 | 0.27 | 0.15 | 0.21 | 1.21 | 1.17 | 0.40 | 0.24 | 0.59 | 3.02 | 0.95 | 0.27 | 0.05 | 0.19 | 0.02 | 0.41 | 0.37 | 0.56 | 3.83 | 6.32 | 0.12 | 0.39 | 17.17 | 12.34 | 0.24 | 1.09 | 85.66 | 9.06 | 0.56 | 0.46 | 0.04 | 0.02 | 0.04 | 0.40 | 0.13 | 0.01 | 0.00 | 0.07 | 0.10 | 0.09 | 2.13 | 0.30 | 4.63 | 292.70 | 9.98 | 0.05 | 5.17 | 0.77 | 20.74 | 38.90 | 0.42 | 8.09 | 4.05 | 0.53 | 6.77 | 14.45 | 13.00 | 8.36 | 6.67 | 30.75 | 0.44 | 0.08 | 0.22 | 2.83 | 3.37 | 9.93 | 2.45 | 1.33 | 1.29 | 10.49 | 23.88 | 20.14 | 0.73 | 0.10 | 0.60 | 2.49 | 2.85 | 35.66 | 11.39 | 34.76 | 0.71 | 0.40 | 4.44 | 43.24 | 8.51 | 14.60 | 6.08 | 10.63 | 6.20 | 0.10 |
| HB_04 | Healthy | 27.80 | 0.67 | 1.67 | 10.33 | 827.38 | 7627.41 | 0.46 | 5.26 | 95.88 | 5715.39 | 90.64 | 18.29 | 92.51 | 0.32 | 0.01 | 0.03 | 0.02 | 0.07 | 1.11 | 0.23 | 0.47 | 0.26 | 2.19 | 2.98 | 3.11 | 0.08 | 0.04 | 0.17 | 0.29 | 0.14 | 0.77 | 0.28 | 0.19 | 1.35 | 0.12 | 1.03 | 1.29 | 3.46 | 1.71 | 5.38 | 5.12 | 1.01 | 0.65 | 5.63 | 2.68 | 0.67 | 3.69 | 5.37 | 1.94 | 0.39 | 0.16 | 7.76 | 0.50 | 0.35 | 0.19 | 0.38 | 0.97 | 0.04 | 0.03 | 0.06 | 0.07 | 0.06 | 0.89 | 1.83 | 8.08 | 2.44 | 0.36 | 214.99 | 67.15 | 130.13 | 0.88 | 0.82 | 0.13 | 0.33 | 0.79 | 0.99 | 5.22 | 116.75 | 1.12 | 0.07 | 0.04 | 9.42 | 2.47 | 0.07 | 1.53 | 0.57 | 1.47 | 0.05 | 0.05 | 0.00 | 0.01 | 0.33 | 0.05 | 0.05 | 0.04 | 2.38 | 5.69 | 2.70 | 3.04 | 1.72 | 0.24 | 0.31 | 0.48 | 0.12 | 0.42 | 0.29 | 0.33 | 0.11 | 0.15 | 0.67 | 0.13 | 14.22 | 34.91 | 8.03 | 0.13 | 3.93 | 10.61 | 6.30 | 557.16 | 103.25 | 2.55 | 0.14 | 13.23 | 16.80 | 5.34 | 3.82 | 0.05 | 171.93 | 522.99 | 207.60 | 66.00 | 363.21 | 34.35 | 0.48 | 0.03 | 32.86 | 6.23 | 0.35 | 69.09 | 117.99 | 2.01 | 0.01 | 10.81 | 1.19 | 0.17 | 32.95 | 8.40 | 5.50 | 5.22 | 2.56 | 0.50 | 17.63 | 6.22 | 1.20 | 5.27 | 53.45 | 3.06 | 29.12 | 46.48 | 3.13 | 10.19 | 1.46 | 2.26 | 0.52 | 5.97 | 10.43 | 2.99 | 20.05 | 1.07 | 7.20 | 11.13 | 2.25 | 0.18 | 7.77 | 0.03 | 0.81 | 1.51 | 0.25 | 0.03 | 0.11 | 0.82 | 3.68 | 0.59 | 2.81 | 0.24 | 0.25 | 7.22 | 2.58 | 3.71 | 0.38 | 0.33 | 0.26 | 0.36 | 0.20 | 1.69 | 0.28 | 7.16 | 4.41 | 4.89 | 0.49 | 0.19 | 0.25 | 1.09 | 1.63 | 0.71 | 0.37 | 1.13 | 3.66 | 1.07 | 0.32 | 0.06 | 0.07 | 0.04 | 0.29 | 0.39 | 0.71 | 5.53 | 9.38 | 0.28 | 0.46 | 21.22 | 12.41 | 0.29 | 1.24 | 90.67 | 7.83 | 0.57 | 0.37 | 0.05 | 0.03 | 0.08 | 0.66 | 0.21 | 0.01 | 0.00 | 0.07 | 0.12 | 0.10 | 2.70 | 0.29 | 4.76 | 388.18 | 12.54 | 0.06 | 5.41 | 0.96 | 30.81 | 37.74 | 0.73 | 9.26 | 4.11 | 0.76 | 9.15 | 23.51 | 13.42 | 13.23 | 9.49 | 43.24 | 0.53 | 0.07 | 0.24 | 4.28 | 1.64 | 5.08 | 1.40 | 0.99 | 2.54 | 8.91 | 15.50 | 9.56 | 0.57 | 0.21 | 0.52 | 1.46 | 6.30 | 22.70 | 6.94 | 16.75 | 0.33 | 1.00 | 7.73 | 21.32 | 3.60 | 5.73 | 2.87 | 6.65 | 3.65 | 0.06 |
| HB_05 | Healthy | 26.21 | 0.93 | 1.76 | 14.02 | 810.18 | 7811.54 | 0.66 | 7.01 | 171.48 | 6238.77 | 50.53 | 18.38 | 90.20 | 0.38 | 0.01 | 0.05 | 0.03 | 0.08 | 1.20 | 0.27 | 0.45 | 0.31 | 2.10 | 3.20 | 3.18 | 0.08 | 0.05 | 0.16 | 0.28 | 0.22 | 0.30 | 0.45 | 0.41 | 2.25 | 0.14 | 1.96 | 2.44 | 4.89 | 2.62 | 10.06 | 11.91 | 1.16 | 0.83 | 4.09 | 3.93 | 0.89 | 3.62 | 4.02 | 1.68 | 0.27 | 0.19 | 4.46 | 0.22 | 0.19 | 0.16 | 0.23 | 0.64 | 0.05 | 0.04 | 0.07 | 0.07 | 0.06 | 0.99 | 1.99 | 8.16 | 2.70 | 0.37 | 205.91 | 69.11 | 137.94 | 0.87 | 0.94 | 0.13 | 0.29 | 0.88 | 1.02 | 7.43 | 126.11 | 0.66 | 0.08 | 0.05 | 8.38 | 2.86 | 0.08 | 2.24 | 0.83 | 2.24 | 0.06 | 0.07 | 0.01 | 0.01 | 0.42 | 0.08 | 0.05 | 0.06 | 2.67 | 6.05 | 2.91 | 3.24 | 1.94 | 0.31 | 0.36 | 0.52 | 0.13 | 0.37 | 0.28 | 0.36 | 0.12 | 0.16 | 0.89 | 0.14 | 17.78 | 27.54 | 6.63 | 0.13 | 3.33 | 6.94 | 6.83 | 618.59 | 73.62 | 2.73 | 0.18 | 12.30 | 16.99 | 4.61 | 4.29 | 0.02 | 143.72 | 456.82 | 205.75 | 71.45 | 360.53 | 30.82 | 0.46 | 0.04 | 31.51 | 7.47 | 0.28 | 100.62 | 115.45 | 1.85 | 0.01 | 13.82 | 1.10 | 0.18 | 34.54 | 9.81 | 5.17 | 9.44 | 4.89 | 0.87 | 20.90 | 6.71 | 1.23 | 7.44 | 81.86 | 5.04 | 40.85 | 62.77 | 3.74 | 13.21 | 2.76 | 3.13 | 0.61 | 8.88 | 10.04 | 4.59 | 24.90 | 0.98 | 10.79 | 12.49 | 2.29 | 0.18 | 4.78 | 0.03 | 0.80 | 1.88 | 0.26 | 0.06 | 0.15 | 0.67 | 3.40 | 0.91 | 2.47 | 0.15 | 0.43 | 7.32 | 2.52 | 5.30 | 0.45 | 0.59 | 0.30 | 0.38 | 0.26 | 1.37 | 0.28 | 9.33 | 4.17 | 5.38 | 0.42 | 0.19 | 0.30 | 1.32 | 1.71 | 0.74 | 0.25 | 0.70 | 4.64 | 0.92 | 0.37 | 0.10 | 0.08 | 0.03 | 0.30 | 0.26 | 0.47 | 3.90 | 5.02 | 0.21 | 0.24 | 8.84 | 10.95 | 0.17 | 0.81 | 86.83 | 7.34 | 0.50 | 0.38 | 0.10 | 0.05 | 0.14 | 1.06 | 0.19 | 0.03 | 0.01 | 0.05 | 0.11 | 0.09 | 2.35 | 0.21 | 5.19 | 437.15 | 11.51 | 0.04 | 5.71 | 1.00 | 36.85 | 50.71 | 0.58 | 12.44 | 4.38 | 0.76 | 8.01 | 23.70 | 15.01 | 11.84 | 10.08 | 47.27 | 0.56 | 0.09 | 0.31 | 7.09 | 3.50 | 9.80 | 2.95 | 1.31 | 3.72 | 16.68 | 29.82 | 19.00 | 1.04 | 0.36 | 0.94 | 2.27 | 13.69 | 40.87 | 11.75 | 25.68 | 0.75 | 2.67 | 14.56 | 36.97 | 7.93 | 9.55 | 3.99 | 6.75 | 3.22 | 0.11 |
9.2 Transform and Scale the Data
Compare this to plotting non-normalized concentrations
# Use broom to make the data ggplot ready and include annotations
pca_annot <- pca_res |> broom::augment(d_long)
pca_contrib <- pca_res |> tidy(matrix = "eigenvalues")
ggplot(data = pca_annot, aes(.fittedPC1, .fittedPC2, color = Group)) +
geom_point(size = 1.5) +
stat_ellipse(level = 0.95) +
theme_light() +
theme(aspect.ratio=1)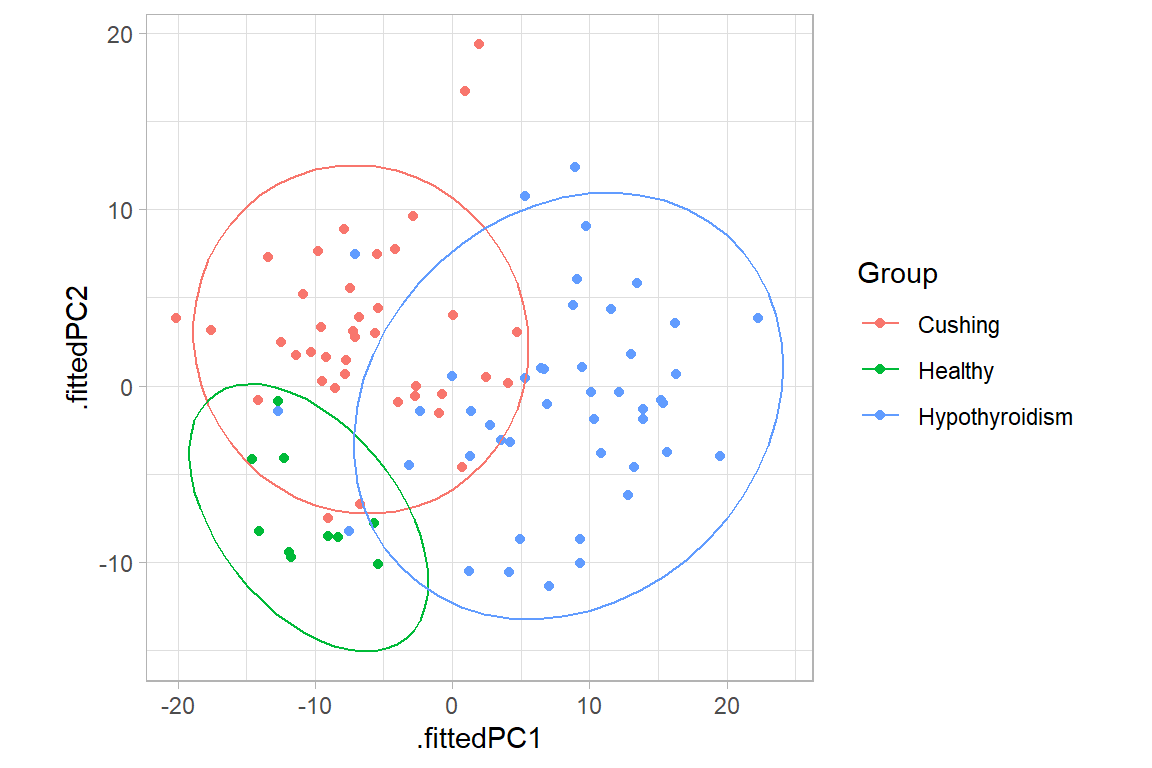
# Use broom to make the data ggplot ready and include annotations
pca_annot <- pca_res |> broom::augment(d_long)
pca_12 <- ggplot(data = pca_annot, aes(.fittedPC1, .fittedPC2, color = Group)) +
geom_point(size = 1.5) +
stat_ellipse(level = 0.95)+
theme_light() +
theme(legend.position = "none")
pca_34 <- ggplot(data = pca_annot, aes(.fittedPC3, .fittedPC4, color = Group)) +
geom_point(size = 1.5) +
stat_ellipse(level = 0.95) +
theme_light() +
theme(legend.position = "none")
cowplot::plot_grid(pca_12, pca_34, nrow = 1,
rel_widths = c(1,1),
labels = c("A","B"))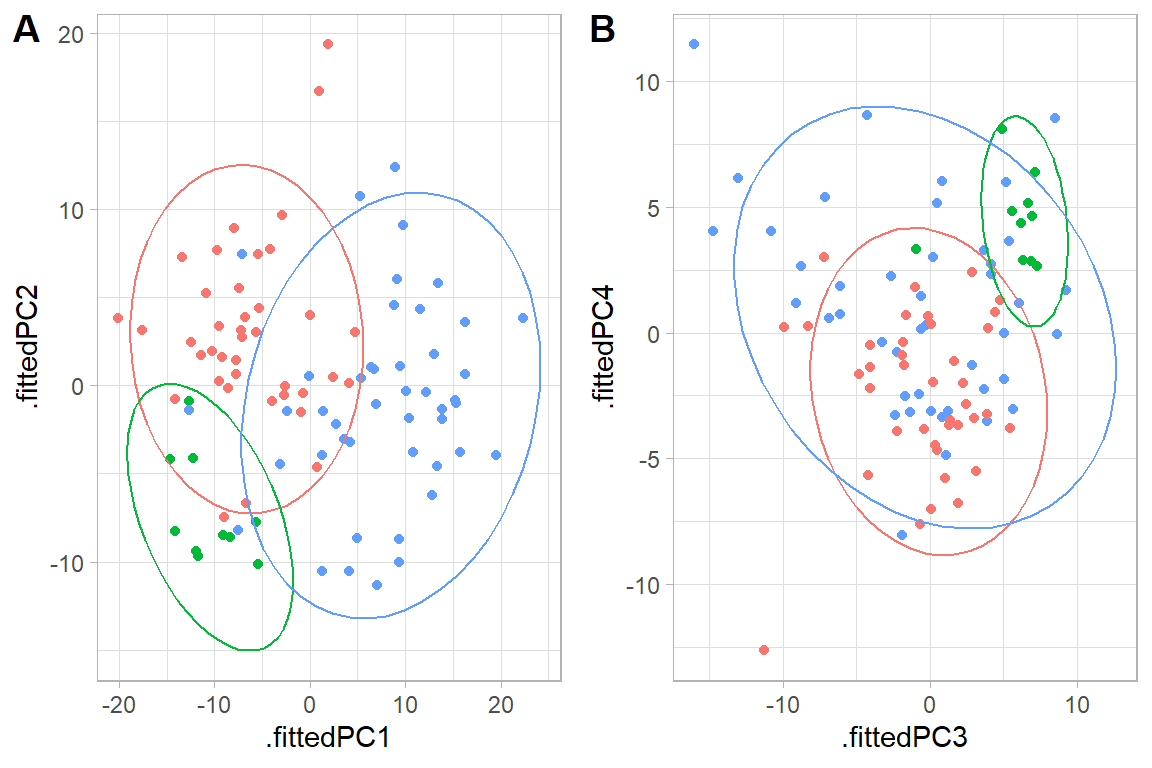
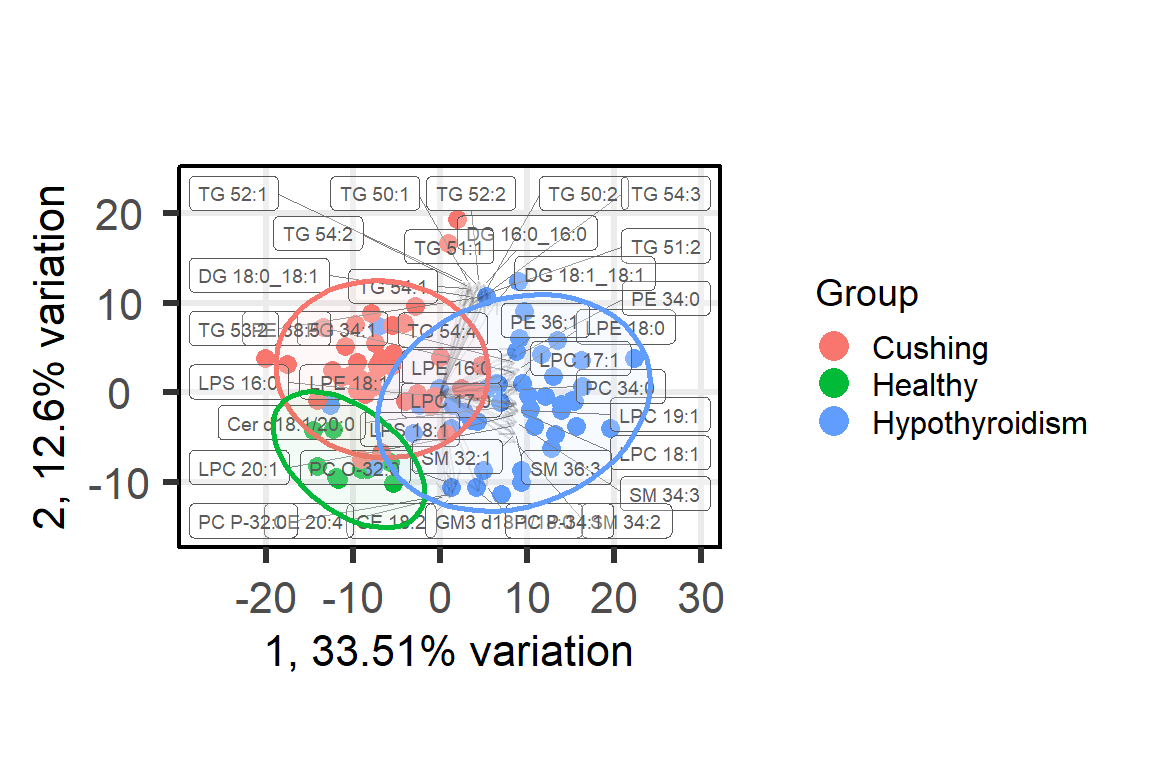
# PCA plot of merged
m_log <- d_long |>
dplyr::select(-Group) |>
dplyr::select(where(~!any(is.na(.)))) |>
column_to_rownames("SubjectID") |>
log2()
d_annot <- d_long |>
select(SubjectID, Group)|>
column_to_rownames("SubjectID")
d_annot$Group <- factor(d_annot$Group)
# get pca result with annotation
pca_res <- PCAtools::pca(mat = m_log, center = TRUE,
metadata = d_annot ,
scale = TRUE,
transposed = TRUE) p_PCA <- PCAtools::biplot(
pca_res,
x = 1,
y = 2 ,
colby = "Group",
#colkey = colby_color,
legendPosition = 'right',
pointSize = 3,
lengthLoadingsArrowsFactor = 1.5,
lab = NULL,
sizeLoadingsNames = 2.5,
alphaLoadingsArrow = 0.1,
colLoadingsNames = "grey35",
showLoadings = TRUE,
ntopLoadings = 20,
drawConnectorsLoadings = T,
boxedLoadingsNames = T,
widthLoadingsArrows = 0.5,
widthConnectorsLoadings = 0.001,
max.overlaps = 33,
ellipseType = "t",
# xlim = c(-22, 25),
# ylim = c(-21, 20),
ellipse = TRUE,
ellipseLevel = 0.95,
ellipseLineSize = 1,
ellipseFill = TRUE,
ellipseAlpha = .05,
gridlines.minor = FALSE,
)
p_PCA
# theme(
# aspect.ratio = 1,
# panel.grid.major = element_line(size = .5),
# legend.position = c(.91, .93),
# legend.title = element_blank(),
# legend.background = element_rect(fill = alpha("white", 0.0), inherit.blank = TRUE)
# )