Create a column which contains a list of ggplot
suited for a pdf report.
Usage
add_ggplot_panel(
curve_table,
curve_summary = NULL,
dilution_table = lifecycle::deprecated(),
dilution_summary = lifecycle::deprecated(),
grouping_variable = c("Curve_Name", "Curve_Batch_Name"),
curve_batch_var = "Curve_Batch_Name",
curve_batch_col = c("#377eb8", "#4daf4a", "#9C27B0", "#BCAAA4", "#FF8A65", "#EFBBCF"),
dil_batch_var = lifecycle::deprecated(),
dil_batch_col = lifecycle::deprecated(),
conc_var = "Concentration",
conc_var_units = "%",
conc_var_interval = 50,
signal_var = "Signal",
have_plot_title = TRUE,
plot_summary_table = TRUE,
plot_first_half_lin_reg = FALSE,
plot_last_half_lin_reg = FALSE
)Arguments
- curve_table
Output given from the function
create_curve_table(). It is in long table format with columns indicating at least the lipid/transition name, the concentration and signal. Other columns may be present if it is used to group the curve together.- curve_summary
The summary table generated by function
summarise_curve_table()and/orevaluate_linearity()but it can also be any generic data frame or tibble. If there is no input given in this, the program will create one using the functionsummarise_curve_table()andevaluate_linearity()withgrouping_variable,conc_varandsignal_varas inputs. Default: NULL- dilution_table
- dilution_summary
- grouping_variable
A character vector of column names in
curve_tableto indicate how each curve should be grouped by. Default: c("Curve_Name", "Curve_Batch_Name")- curve_batch_var
Column name in
curve_tableto indicate the group name of each curve batch, used to colour the points in the curve plot. Default: 'Curve_Batch_Name'- curve_batch_col
A vector of colours to be used for the curve batch group named given in
curve_batch_var. Default: c("#377eb8", "#4daf4a", "#9C27B0", "#BCAAA4", "#FF8A65", "#EFBBCF")- dil_batch_var
- dil_batch_col
- conc_var
Column name in
curve_tableto indicate concentration. Default: 'Concentration'- conc_var_units
Unit of measure for
conc_var. Default: '%'- conc_var_interval
Distance between two tick labels in the curve plot. Default: 50
- signal_var
Column name in
curve_tableto indicate signal. Default: 'Area'- have_plot_title
Indicate if you want to have a plot title in the
ggplotplot. Default: TRUE- plot_summary_table
Indicate if you want to plot the summary table in the
ggplotplot. Default: TRUE- plot_first_half_lin_reg
Decide if we plot an extra regression line that best fits the first half of
conc_varcurve points. Default: FALSE- plot_last_half_lin_reg
Decide if we plot an extra regression line that best fits the last half of
conc_varcurve points. Default: FALSE
Value
A table with columns from grouping variable
and a new column panel created containing a ggplot curve plot
in each row. This column is used to create the plot figure in the
pdf report.
Examples
# Data Creation
concentration <- c(
10, 20, 25, 40, 50, 60,
75, 80, 100, 125, 150,
10, 25, 40, 50, 60,
75, 80, 100, 125, 150
)
curve_batch_name <- c(
"B1", "B1", "B1", "B1", "B1",
"B1", "B1", "B1", "B1", "B1", "B1",
"B2", "B2", "B2", "B2", "B2",
"B2", "B2", "B2", "B2", "B2"
)
sample_name <- c(
"Sample_010a", "Sample_020a",
"Sample_025a", "Sample_040a", "Sample_050a",
"Sample_060a", "Sample_075a", "Sample_080a",
"Sample_100a", "Sample_125a", "Sample_150a",
"Sample_010b", "Sample_025b",
"Sample_040b", "Sample_050b", "Sample_060b",
"Sample_075b", "Sample_080b", "Sample_100b",
"Sample_125b", "Sample_150b"
)
curve_1_saturation_regime <- c(
5748124, 16616414, 21702718, 36191617,
49324541, 55618266, 66947588, 74964771,
75438063, 91770737, 94692060,
5192648, 16594991, 32507833, 46499896,
55388856, 62505210, 62778078, 72158161,
78044338, 86158414
)
curve_2_good_linearity <- c(
31538, 53709, 69990, 101977, 146436, 180960,
232881, 283780, 298289, 344519, 430432,
25463, 63387, 90624, 131274, 138069,
205353, 202407, 260205, 292257, 367924
)
curve_3_noise_regime <- c(
544, 397, 829, 1437, 1808, 2231,
3343, 2915, 5268, 8031, 11045,
500, 903, 1267, 2031, 2100,
3563, 4500, 5300, 8500, 10430
)
curve_4_poor_linearity <- c(
380519, 485372, 478770, 474467, 531640, 576301,
501068, 550201, 515110, 499543, 474745,
197417, 322846, 478398, 423174, 418577,
426089, 413292, 450190, 415309, 457618
)
curve_batch_annot <- tibble::tibble(
Sample_Name = sample_name,
Curve_Batch_Name = curve_batch_name,
Concentration = concentration
)
curve_data <- tibble::tibble(
Sample_Name = sample_name,
`Curve_1` = curve_1_saturation_regime,
`Curve_2` = curve_2_good_linearity,
`Curve_3` = curve_3_noise_regime,
`Curve_4` = curve_4_poor_linearity
)
# Create curve table
curve_table <- create_curve_table(
curve_batch_annot = curve_batch_annot,
curve_data_wide = curve_data,
common_column = "Sample_Name",
signal_var = "Signal",
column_group = "Curve_Name"
)
# Create curve statistical summary
curve_summary <- curve_table |>
summarise_curve_table(
grouping_variable = c(
"Curve_Name",
"Curve_Batch_Name"
),
conc_var = "Concentration",
signal_var = "Signal"
) |>
dplyr::arrange(.data[["Curve_Name"]]) |>
evaluate_linearity(grouping_variable = c(
"Curve_Name",
"Curve_Batch_Name"
))
# Create a ggplot table
ggplot_table <- add_ggplot_panel(
curve_table,
curve_summary = curve_summary,
grouping_variable = c("Curve_Name",
"Curve_Batch_Name"),
curve_batch_var = "Curve_Batch_Name",
conc_var = "Concentration",
conc_var_units = "%",
conc_var_interval = 50,
signal_var = "Signal"
)
ggplot_list <- ggplot_table$panel
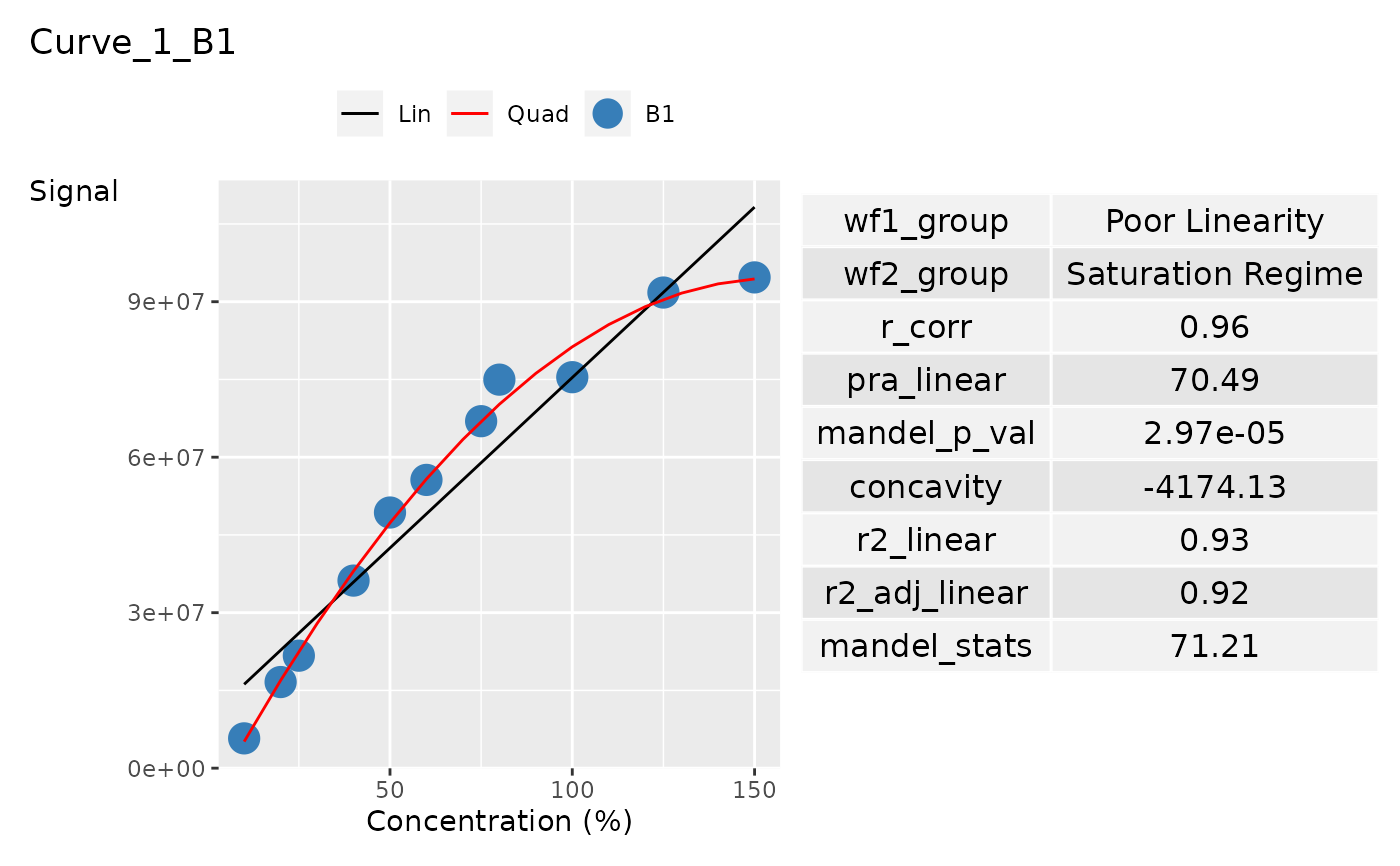
ggplot_list[[1]]
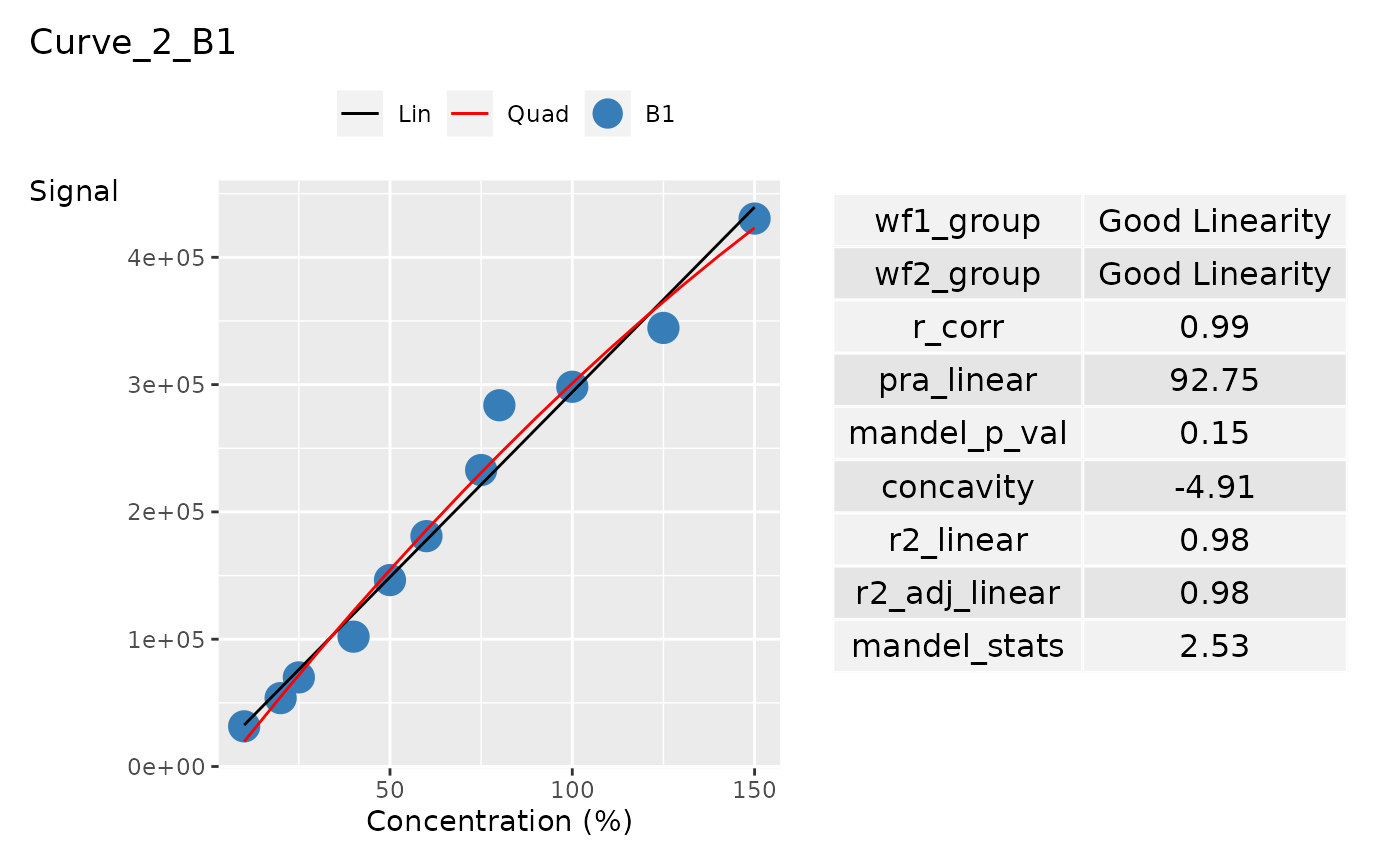
 ggplot_list[[2]]
ggplot_list[[2]]
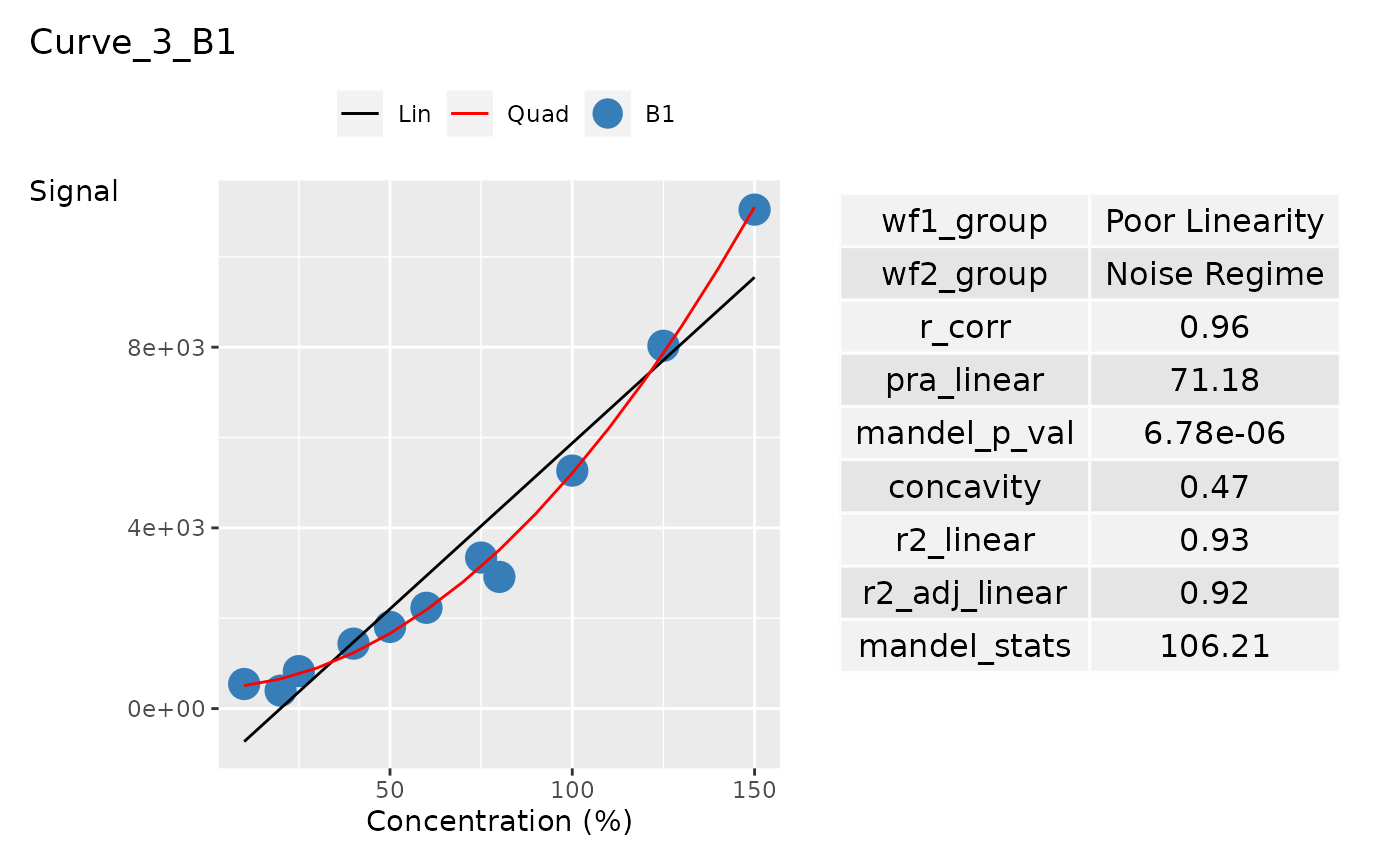
 ggplot_list[[3]]
ggplot_list[[3]]